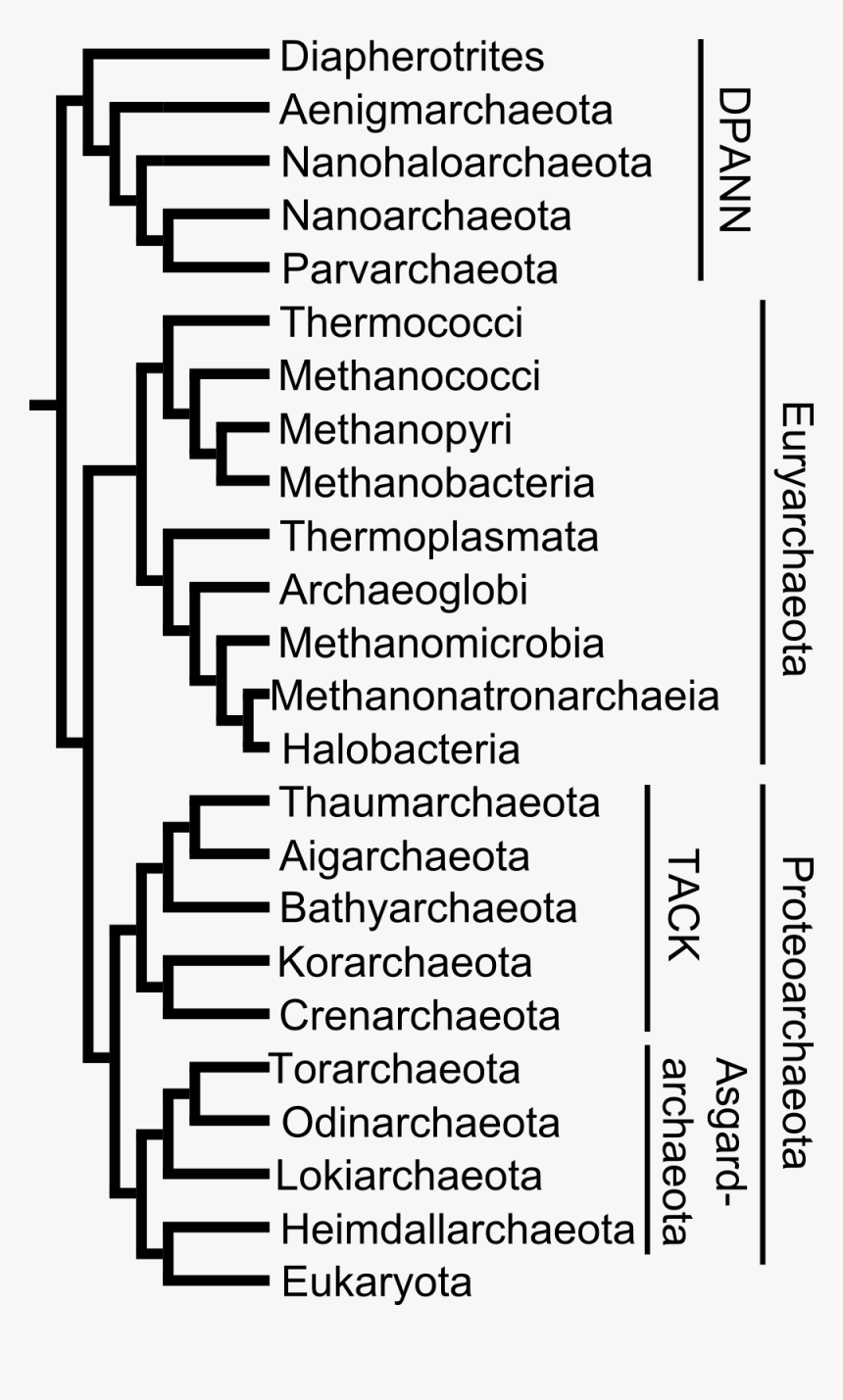
Aquaporins (AQPs) are membrane channel proteins found in a wide range of organisms, from archaea and bacteria to plants and animals. AQPs facilitate the rapid transport of water across cellular membranes and are of fundamental importance to the control of cell volume and transcellular water traffic. The Microbe Directory is a collective research effort to profile and annotate more than 7,500 unique microbial species from the MetaPhlAn2 database that includes bacteria, archaea, viruses, fungi, and protozoa. By collecting and summarizing data on various microbes' characteristics, the project comprises a database that can be used downstream of large-scale metagenomic taxonomic analyses.
August 10, 2013. Softberry have developed gene finding parameters for 30 new organisms to support gene-prediction program suit and next-generation sequencing data analysis (Transomics pipeline to discover alternatively spliced gene variants).
December 24, 2012. Softberry provided download about 100 software applications for free usage in research academic project. The download WEB page can be viewed here.
October 28, 2011. Softberry announces 11th release of RegSite database of plant regulatory elements. A list of its 2,252 entries representing 2,286 motifs can be viewed here.
The graphical user interface (GUI) is a native Microsoft Windows application that can also be used on Mac OS X. The command line M ega is available as native applications for Windows, Linux,.
October 15, 2010. Softberry completes development of cross-platform version of MolQuest, the most comprehensive desktop application for sequence analysis. MolQuest contains more than 100 programs, including those for gene and promoter finding, regulatory elements mapping, pattern discovery, multiple sequences alignment, phylogenetic reconstruction, protein structure predictions and various statistical data analysis and many others, as well as dozens of viewers and a tool for building custom pipelines. Fully functional trial version of MolQuest for Windows, Mac OS X or Linux can be downloaded at www.molquest.com.
October 14, 2009. Softberry announces a pipeline for assembly and analysis of transcriptome data obtained using next generation sequencing technologies. The pipeline can be used to estimate gene expression levels, identify alternative splice variants, and improve gene proediction and genome annotation using transcription data. More details are available here.
October 9, 2009. Release 7 of RegSite plant regulatory elements database is available. The release contains 1816 antries, a list of which can be seen here.
February 14, 2008. Softberry releases OligoZip package for analysis of Solexa/Illumina and similar sequencing reads. It can be used for genomic sequence assembly - de novo or based on a reference genome - and mutation profiling ans SNP discovery.
January 31, 2008. Softberry releases MolQuest 2.0 Alpha version, which supports user-customizable pipelines. Its trial version can be downloaded at www.molquest.com
August 28, 2007. FGENESH++ pipeline was a winner of NGASP Gene prediction competition on exon level (see NGASP Gene prediction competition Results), while both FGENESH and FGENESH++ had best sensitivity in most categories. See here for NGASP comparison of various genefinders.
June 28, 2007. Independent tests of accuracy of ORF prediction by FGENESB on metagenomic sequences found it to be far superior to alternative methods, such as Critica and Glimmer (Mavromatis et al., NATURE METHODS, 2007, Vol. 4 No. 6, p. 495-500), see here for details.
December 13, 2006. Softberry releases parameters for honey bee, grape (Vitis vinifera), tomato, Bos taurus, Medicago, and several new fungal genomes. New parameters can be used with FGENESH, FEX and BESTORF (here) and FGENESH+, FGENESH_C and FGENESH-2 programs (here). The accuracy with FGENESH is 93-97% on nucleotide level.
December 11, 2006. Snow roller mac os. Fifth release of RegSite database of plant regulatory motifs is now available. RegSite V.5 contains 1211 entries and more descriptive fields than other plant motifs databases. A list of RegSite entries is available here. To search for plant functional motifs from updated RegSite DB using NSITE-PL program, click here.
Jul 25, 2006. MolQuest ver. 1.5 is released. This version contains possibility to build and execute simple pipelines as well as improving interfaces in many viewers as well as correction some programs such as FoldRNA (which worked in best palindrome mode earlier). Trial version of Molquest can be downloaded here.
June 30, 2006. MolQuest ver. 1.4 is released, which includes 19 new programs for DNA, RNA and protein sequence analysis and five graphical tools for visualization of programs output. Trial version of Molquest for Windows can be downloaded here.
March 21, 2006. Softberry releases two protein 3D structure analysis programs. 3D-Match aligns 3D structures of two proteins, while 3D-MatchDB performs fast search of nonredundant PDB for structural homology with a query protein. Both programs can be tried here
Humanly optimized mac os. December 20, 2005. Fourth release of RegSite database of plant regulatory motifs is now available. RegSite V.4 contains 1009 entries and more descriptive fields than other plant motifs databases. A list of RegSite entries is available here. To search for plant functional motifs from updated RegSite DB using NSITE-PL program, click here.
February 18, 2005. Softberry releases genefinding parameters for 240 prokaryotic genomes, including 25 archaea. Parameters are available for use with FGENESB bacterial gene and operon finding program here. Besides genome-specific parameters, they include generic eubacterial, archaea and mixed eubacterial-archaea parameters suitable for analysis of bacterial communities.
February 11, 2005. Softberry releases genefinding parameters for chicken and Xenopus New parameters can be used with FGENESH, FEX and BESTORF (here) and FGENESH+, FGENESH_C and FGENESH-2 programs (here). The accuracy with FGENESH is 95-97% on nucleotide level.
October 4, 2004. Softberry releases ProtComp ver. 6. The new version of popular program for prediction of protein subcellular localization, ProtComp, has overall prediction accuracy of >90% (see here for more details). Prediction accuracy of prokaryotic version, ProtCompB ver. 2, is 95%. Both programs can be tried here.
September 23, 2004. Softberry posts gene finding parameters for ten new genomes. These paramenets can be used for gene prediction in those and related genomes with FGENESH, FGENESH+, FGENESH_C, FGENESH-2, FEX and BESTORF programs.
May 18, 2004. Softberry releases 3D-Explorer: A powerful viewer of spatial models of biological macromolecules and their complexes. Macromolecule can be presented as wire, stick, ball and stick, and CPK models. Charge or hydrophobicity of molecular surface can also be visualized. 3D-Explorer reads PDB files, is compatible with GetAtoms and CE applications and can show two superimposed 3D structures. Trial version of 3D-Exporer can be downloaded here.
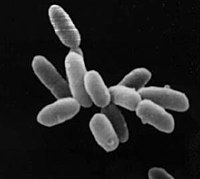
Archaea Terata Mac Os Catalina
May 14, 2004. Softberry introduces a new program CYS_REC. The program performs the first step in locating of disulphide briges in proteins: prediction of SS-bonding states of cysteines. It can be useful in construction of protein 3D structures. CYS_REC can be tried here.
May 5, 2004. Softberry introduces protein-to-genome mapping program, Prot_Map. The program maps a set of protein sequences onto genome, producing gene structures and alignments of coding exons with proteins. The program is many times faster than Genewise and does not require knowledge of protein genomic location. Gene structures predicted by Prot_Map may be further refined by FGENESH+, producing very accurate gene models.
Prot_map program can be used (1) as additional element of FGENESH++C automatic genomic sequence annotation pipeline, (2) to generate a set of predicted genes to train parameters for gene finding programs like FGENESH on newly sequenced genomes, where little other information is available, and (3) to find pseudogenes by selecting corrupted gene structures found by mapping a set of known proteins. Prot_Map can be accessed here.
March 21, 2005. Softberry releases two protein 3D structure analysis programs. 3D-Match aligns 3D structures of two proteins, while 3D-MatchDB performs fast search of nonredundant PDB for structural homology with a query protein. Both programs can be tried here
December 20, 2005. Fourth release of RegSite database of plant regulatory motifs is now available. RegSite V.4 contains 1009 entries and more descriptive fields than other plant motifs databases. A list of RegSite entries is available here. To search for plant functional motifs from updated RegSite DB using NSITE-PL program, click here.
February 18, 2005. Softberry releases genefinding parameters for 240 prokaryotic genomes, including 25 archaea. Parameters are available for use with FGENESB bacterial gene and operon finding program here. Besides genome-specific parameters, they include generic eubacterial, archaea and mixed eubacterial-archaea parameters suitable for analysis of bacterial communities.
February 11, 2005. Softberry releases genefinding parameters for chicken and Xenopus New parameters can be used with FGENESH, FEX and BESTORF (here) and FGENESH+, FGENESH_C and FGENESH-2 programs (here). The accuracy of new parameters with FGENESH is 95-97% on nucleotide level.
September 11, 2003. Softberry develops genefinding parameters for fish and algae New parameters can be used with all genefinders of FGENESH suite and also FEX and BESTORF programs. Their accuracy of coding region identification with FGENESH is about 98% on nucleotide level. New parameters can be tried here.
August 19, 2003. RegSite-Plant Database (V.2) of regulatory motifs is released. RegSite-Plant contains 702 entries and more descriptive fields than the other plant motifs databases including several fields on expression and some others. To see Database content and format or predict plant promoter sequences and analize regulatory motifs, click here.
Archaea Terata Mac Os X
July 30, 2003. Softberry introduces PDISORDER ver. 1.0, the program for predicting ordered and disordered regions in protein sequences. Intrinsically unstructured (disordered) protein regions play key roles in cell-signaling, regulation and cancer, making them extremely useful for discovery of anticancer drugs. PDISORDER can be tried here.
May 12, 2003. ProtComp ver. 5 is released. For this release, we retrained neural networks on much larger sample of proteins with known localization, which resulted in about 10% increase in accuracy of protein localization prediction. PortComp can be tried here.
April 15, 2003. Softberry annotates SARS associated coronavirus. Softberry releases annotation of SARS associated coronavirus TOR2 with FGENESV viral gene finder and FGENESV-Annotator script. To access annotation, click here.
March 19, 2003. PromH, mammal promoter finding program with improved accuracy, is released. To further improve promoter identification accuracy achieved by Softberry's TSSW and TSSG programs, we developed a new pair of programs that takes into account conserved features of major promoter functional components, such as transcription start points, TATA-boxes and regulatory motifs, in pairs of orthologous genes aligned by SCAN2 program, which results in significant accuracy improvement. PromH(G) variant uses elements of TFD database, while promH(W) variant - those of TRANSFAC® database. Both programs can be tried here.
January 20, 2003. Softberry introduces BPROM, bacterial promoter finding program. The program, which predicts bacterial sigma70 promoters with about 80% accuracy and is a useful tool for improving bacterial operon prediction, can be tried at Softberry promoter finding or bacterial gene finding pages.
January 9, 2003. Softberry annotates rat (Rattus norvegicus) genome. The annotation can be viewed in Softberry or at UCSC Genome Bioinformatics Group site in SC Genome Browser.
December 19, 2002. Softberry introduces gene finding parameters for Nicotiana tabacum. Accuracy of ab initio gene prediction on tobacco genome is about 98% on nucleotide level. Parameters for tobacco gene prediction can be tried with FGENESH here.
November 19, 2002. Softberry annotates genomes of several pathogenic bacteria. Using FGENESB-Annotator script, Softberry has annotated genomes of Mycobacterium tuberculosis strains H37Rv and CDC1551, and Yersinia pestis CO92 and KIM, in addition to annotation of main chromosome of Bacillus anthracis completed earlier.
November 5, 2002. Softberry introduces gene finding parameters for Anopheles gambiae, with accuracy of ab initio gene prediction about 98% on nucleotide level. These and nine other available parameters can be tried with FGENESH here.
October 31, 2002. Gene finding parameters for Plasmodium falciparum are now available. These parameters, developed for FGENESH family of programs, provide 98% accuracy on nucleotide level and 80% accuracy of exact ab initio exon prediction. You can try FGENESH with these and eight other available parameters here.
October 23, 2002. Softberry introduces FGENESV, gene prediction program for viral genomes. The program is based on pattern recognition of different types of signals and Markov chain models of coding regions. Optimal combination of these features is then found by dynamic programming and a set of gene models is constructed along given sequence. FGENESV can be trained on genome sequence of query virus, or used with generic parameters (FGENESV0 variant) on small genomes which do not have enough statistics for parameter training. Both trainable and generic variants can be tried here.
March 21, 2005. Softberry releases two protein 3D structure analysis programs. 3D-Match aligns 3D structures of two proteins, while 3D-MatchDB performs fast search of nonredundant PDB for structural homology with a query protein. Both programs can be tried here
December 20, 2005. Fourth release of RegSite database of plant regulatory motifs is now available. RegSite V.4 contains 1009 entries and more descriptive fields than other plant motifs databases. A list of RegSite entries is available here. To search for plant functional motifs from updated RegSite DB using NSITE-PL program, click here.
February 18, 2005. Softberry releases genefinding parameters for 240 prokaryotic genomes, including 25 archaea. Parameters are available for use with FGENESB bacterial gene and operon finding program here. Besides genome-specific parameters, they include generic eubacterial, archaea and mixed eubacterial-archaea parameters suitable for analysis of bacterial communities.
February 11, 2005. Softberry releases genefinding parameters for chicken and Xenopus New parameters can be used with FGENESH, FEX and BESTORF (here) and FGENESH+, FGENESH_C and FGENESH-2 programs (here). The accuracy of new parameters with FGENESH is 95-97% on nucleotide level.
September 11, 2003. Softberry develops genefinding parameters for fish and algae New parameters can be used with all genefinders of FGENESH suite and also FEX and BESTORF programs. Their accuracy of coding region identification with FGENESH is about 98% on nucleotide level. New parameters can be tried here.
August 19, 2003. RegSite-Plant Database (V.2) of regulatory motifs is released. RegSite-Plant contains 702 entries and more descriptive fields than the other plant motifs databases including several fields on expression and some others. To see Database content and format or predict plant promoter sequences and analize regulatory motifs, click here.
Archaea Terata Mac Os X
July 30, 2003. Softberry introduces PDISORDER ver. 1.0, the program for predicting ordered and disordered regions in protein sequences. Intrinsically unstructured (disordered) protein regions play key roles in cell-signaling, regulation and cancer, making them extremely useful for discovery of anticancer drugs. PDISORDER can be tried here.
May 12, 2003. ProtComp ver. 5 is released. For this release, we retrained neural networks on much larger sample of proteins with known localization, which resulted in about 10% increase in accuracy of protein localization prediction. PortComp can be tried here.
April 15, 2003. Softberry annotates SARS associated coronavirus. Softberry releases annotation of SARS associated coronavirus TOR2 with FGENESV viral gene finder and FGENESV-Annotator script. To access annotation, click here.
March 19, 2003. PromH, mammal promoter finding program with improved accuracy, is released. To further improve promoter identification accuracy achieved by Softberry's TSSW and TSSG programs, we developed a new pair of programs that takes into account conserved features of major promoter functional components, such as transcription start points, TATA-boxes and regulatory motifs, in pairs of orthologous genes aligned by SCAN2 program, which results in significant accuracy improvement. PromH(G) variant uses elements of TFD database, while promH(W) variant - those of TRANSFAC® database. Both programs can be tried here.
January 20, 2003. Softberry introduces BPROM, bacterial promoter finding program. The program, which predicts bacterial sigma70 promoters with about 80% accuracy and is a useful tool for improving bacterial operon prediction, can be tried at Softberry promoter finding or bacterial gene finding pages.
January 9, 2003. Softberry annotates rat (Rattus norvegicus) genome. The annotation can be viewed in Softberry or at UCSC Genome Bioinformatics Group site in SC Genome Browser.
December 19, 2002. Softberry introduces gene finding parameters for Nicotiana tabacum. Accuracy of ab initio gene prediction on tobacco genome is about 98% on nucleotide level. Parameters for tobacco gene prediction can be tried with FGENESH here.
November 19, 2002. Softberry annotates genomes of several pathogenic bacteria. Using FGENESB-Annotator script, Softberry has annotated genomes of Mycobacterium tuberculosis strains H37Rv and CDC1551, and Yersinia pestis CO92 and KIM, in addition to annotation of main chromosome of Bacillus anthracis completed earlier.
November 5, 2002. Softberry introduces gene finding parameters for Anopheles gambiae, with accuracy of ab initio gene prediction about 98% on nucleotide level. These and nine other available parameters can be tried with FGENESH here.
October 31, 2002. Gene finding parameters for Plasmodium falciparum are now available. These parameters, developed for FGENESH family of programs, provide 98% accuracy on nucleotide level and 80% accuracy of exact ab initio exon prediction. You can try FGENESH with these and eight other available parameters here.
October 23, 2002. Softberry introduces FGENESV, gene prediction program for viral genomes. The program is based on pattern recognition of different types of signals and Markov chain models of coding regions. Optimal combination of these features is then found by dynamic programming and a set of gene models is constructed along given sequence. FGENESV can be trained on genome sequence of query virus, or used with generic parameters (FGENESV0 variant) on small genomes which do not have enough statistics for parameter training. Both trainable and generic variants can be tried here.
September 4, 2002. Softberry includes simple operon recognition in its FGENESB gene prediction program, releases FGENESB-Annotator, an advanced bacterial gene predictor, and annotates Bacillus anthracis genome. New FGENESB version, which can be tried here, now includes simple operon prediction model based on gene distances. Increasing accuracy of operon identification using promoter, terminator and other features is under development. We have also developed new FGENESB-Annotator script that finds similar proteins in public databases and annotates predicted bacterial genes. This script can also identify low scoring genes if they have known homologous protein. Using FGENESB-Annotator script, we annotated Bacillus anthracis A2012 main chromosome and thirteen other sequenced bacterial genomes.
August 27, 2002. Softberry releases bacterial genefinder, FGENESB - the latest addition to its family of gene finding programs. Like other Softberry gene finders, FGENESB is the fastest (E.coli genome is annotated in ~14 sec) and most accurate ab initio bacterial gene prediction program available. It takes only ten minutes to train the program on new genome. FGENESB is based on pattern recognition of different types of signals and Markov chain models of coding regions. You can try it here.
July 10, 2002. TSSP, the program for finding plant promoters and transcription start points, is released. TSSP uses functional motifs selected from RegSite database developed by Softberry. You can try TSSP here .
June 27, 2002. NSITE-PL, the program for finding statisticaly significant functional motifs in plant promoter/regulatory sequences, is released. Plant functional motifs are selected from RegSite database developed by Softberry Inc. from published data on transcription regulation of plant genes. You can try NSITE-PL here. RegSite DB is also available for licensing.
April 19, 2002. Softberry introduces the first human-mouse syntheny alignment based on draft genomic sequences. Compared to NCBI homology maps, Softberry map contains significantly more genes (14504 ortologous pairs) and is directly linked to genomic sequences. The data were generated by Softberry programs for gene prediction, EST/RNA mapping and gemomic sequence comparison - some of them can be tested at this site. To access syntheny information, click here.
April 19, 2002. Softberry FGENESH gene prediction program is cited as the best annotation tool in two rice genome sequencing projects, as published in April 5, 2002 issue of Science. 'For rice, however, FGeneSH is the most successful (gene finding) program', acknowleges a group led by Beijing Genomics Institute. Syngenta-led project selected 87% of high-evidence genes based on FGENESH output.
April 1, 2002. GETATOMS - The program for modeling atomic coordinates of a protein with unknown 3D structure. GETATOMS uses main chain coordinates from 3D structure of similar protein, which sequence is aligned with a query protein. Restoration of loops in alignment will be added in later versions. GETATOMS also has an option to provide coordinates of H-atoms. You can try it here.
Archaea Terata Mac Os 11
March 22, 2002.Softberry completes FGENESH++ annotation of mouse genome draft. You can search complete databases of mouse predicted exons and proteins for homology with your query sequence here
March 21, 2002.Softberry selects CTC Laboratory Systems Corporation as exclusive distributor of its software in Japan. Softberry, Inc. and CTC Laboratory Systems Corporation (CTCLS) announce the strategic agreement granting CTCLS exclusive rights to distribute Softberry's entire product line in Japan. CTCLS will also provide front-line maintenance for Softberry's products at that market. More

